Bay, Ömer Faruk
Loading...

Profile URL
Name Variants
Bay, Omer F.
Job Title
Öğr. Gör.
Email Address
omerfaruk.bay@agu.edu.tr
Main Affiliation
04.03. Biyoenformatik
Status
Current Staff
Website
ORCID ID
Scopus Author ID
Turkish CoHE Profile ID
Google Scholar ID
WoS Researcher ID
Sustainable Development Goals
SDG data is not available

This researcher does not have a Scopus ID.

This researcher does not have a WoS ID.

Scholarly Output
1
Articles
1
Views / Downloads
5/0
Supervised MSc Theses
0
Supervised PhD Theses
0
WoS Citation Count
5
Scopus Citation Count
5
WoS h-index
1
Scopus h-index
1
Patents
0
Projects
0
WoS Citations per Publication
5.00
Scopus Citations per Publication
5.00
Open Access Source
1
Supervised Theses
0
Google Analytics Visitor Traffic
| Journal | Count |
|---|---|
| Nature Communications | 1 |
Current Page: 1 / 1
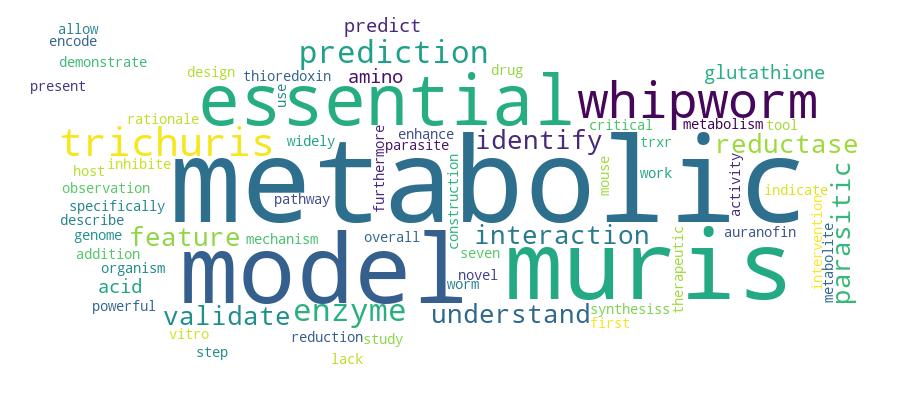
Competency Cloud